Download data#
You can compose queries to download raw data.
import doplaydo.dodata as dd
import matplotlib.pyplot as plt
import pandas as pd
import getpass
username = getpass.getuser()
PROJECT_ID = f"spirals-{username}"
You have access to:
dd.Project
dd.Die
dd.Wafer
dd.ParentCell
dd.Cell
dd.Device
dd.DeviceData
Where each model has its table columns as attributes.

You can use get_data_by_query to query a subset of data filtered by a list of clauses.
It will return a list of tuples where the first element is a DeviceData object and the second one is a pandas DataFrame.
data_tuples = dd.get_data_by_query(
[
dd.Project.project_id == PROJECT_ID,
],
limit=1,
)
device_data, df = data_tuples[0] # each tuple has DeviceData and pd.Dataframe
device_data
DeviceData(pkey=7215, data_type=<DeviceDataType.measurement: 'measurement'>, thumbnail_path='device_data/7215/thumbnail.png', plotting_kwargs={'x_col': 'wavelength', 'y_col': ['output_power'], 'x_name': 'wavelength', 'y_name': 'output_power', 'scatter': False, 'sort_by': {}, 'x_units': None, 'y_units': None, 'grouping': {}, 'x_limits': None, 'y_limits': None, 'x_log_axis': False, 'y_log_axis': False}, valid=True, device_pkey=609, path='device_data/7215/data.json.gz', attributes={}, die_pkey=795, timestamp=datetime.datetime(2025, 2, 28, 21, 4, 41, 320711), timestamp_acquired=None)
device_id = device_data.device.device_id
device_id
'RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_20150_60150'
df
| wavelength | output_power | polyfit | |
|---|---|---|---|
| 0 | 1500.000000 | 0.034341 | 6.0 |
| 1 | 1500.833333 | 0.034941 | 6.0 |
| 2 | 1501.666667 | 0.036901 | 6.0 |
| 3 | 1502.500000 | 0.042276 | 6.0 |
| 4 | 1503.333333 | 0.039977 | 6.0 |
| ... | ... | ... | ... |
| 116 | 1596.666667 | 0.041897 | 6.0 |
| 117 | 1597.500000 | 0.039914 | 6.0 |
| 118 | 1598.333333 | 0.039522 | 6.0 |
| 119 | 1599.166667 | 0.039557 | 6.0 |
| 120 | 1600.000000 | 0.029834 | 6.0 |
121 rows × 3 columns
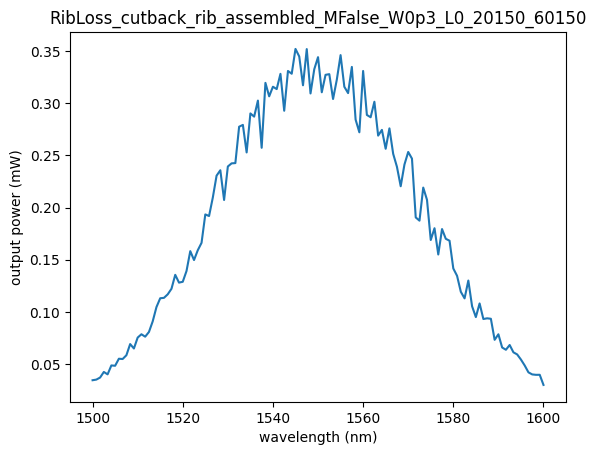
plt.plot(df["wavelength"], df["output_power"])
plt.xlabel("wavelength (nm)")
plt.ylabel("output power (mW)")
plt.title(device_id)
Text(0.5, 1.0, 'RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_20150_60150')
You can aggregate the dataframes and the device data objects
dfs = [dt[1] for dt in data_tuples] # dataframes
dds = [dt[0] for dt in data_tuples] # device data objects
You can use the DeviceData object to traverse the data model and access additional fields.
You can go from DeviceData to any other tables by following the dashed arrows.
Each column is an attribute on the object representing the table.

print("device name: ", dds[0].device.device_id)
print("die x: ", dds[0].die.x)
print("die y: ", dds[0].die.y)
print("wafer name: ", dds[0].die.wafer.wafer_id)
print("cell name: ", dds[0].device.cell.cell_id)
print("parent cell name: ", dds[0].device.parent_cell.cell_id)
print("project name: ", dds[0].device.cell.project.project_id)
device name: RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_20150_60150
die x: 0
die y: 2
wafer name: 6d4c615ff105
cell name: cutback_rib_assembled_MFalse_W0p3_L0
parent cell name: RibLoss
project name: spirals-runner
For example, you can reach the Cell table and all its columns:
dds[0].device.cell
Cell(attributes={'x': 20150, 'y': 60150, 'width_um': 0.3, 'length_um': 0, 'kfactory:ports:0': "{'cross_section': '1da116c1_100', 'info': {}, 'name': 'cutback_rib_W0p3_L0_243700_-2000_ALIGN_IN', 'port_type': 'fiber_launch', 'trans': r270 0,-24}", 'kfactory:ports:1': "{'cross_section': '1da116c1_100', 'info': {}, 'name': 'cutback_rib_W0p3_L0_243700_-2000_ALIGN_OUT', 'port_type': 'fiber_launch', 'trans': r270 150000,-24}", 'kfactory:ports:2': "{'cross_section': '1da116c1_100', 'info': {}, 'name': 'cutback_rib_W0p3_L0_243700_-2000_IN', 'port_type': 'fiber_launch', 'trans': r270 50000,-24}", 'kfactory:ports:3': "{'cross_section': '1da116c1_100', 'info': {}, 'name': 'cutback_rib_W0p3_L0_243700_-2000_OUT', 'port_type': 'fiber_launch', 'trans': r270 100000,-24}", 'kfactory:settings': "{'length': 0, 'mirror': False, 'width': 0.30000000000000004}", 'kfactory:function_name': 'cutback_rib_assembled'}, timestamp=datetime.datetime(2025, 2, 28, 21, 4, 4, 847248), pkey=610, cell_id='cutback_rib_assembled_MFalse_W0p3_L0', project_pkey=21)
Build table for JMP#
You can build a flat table by combining all the device data and pandas dataframes.
data_tuples = dd.get_data_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.Device.device_id
== "RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_20150_60150",
dd.Die.x == 0,
dd.Die.y == 0,
]
)
len(data_tuples)
dds = [dt[0] for dt in data_tuples] # device data objects
dfs = [dt[1] for dt in data_tuples] # dataframes
dfs[0]
| wavelength | output_power | polyfit | |
|---|---|---|---|
| 0 | 1500.000000 | 0.031264 | 6.0 |
| 1 | 1500.833333 | 0.031813 | 6.0 |
| 2 | 1501.666667 | 0.036398 | 6.0 |
| 3 | 1502.500000 | 0.044324 | 6.0 |
| 4 | 1503.333333 | 0.039984 | 6.0 |
| ... | ... | ... | ... |
| 116 | 1596.666667 | 0.041625 | 6.0 |
| 117 | 1597.500000 | 0.043175 | 6.0 |
| 118 | 1598.333333 | 0.036865 | 6.0 |
| 119 | 1599.166667 | 0.034557 | 6.0 |
| 120 | 1600.000000 | 0.032933 | 6.0 |
121 rows × 3 columns
dfs_all = []
for device_data, df in zip(dds, dfs):
df["device_id"] = device_data.device.device_id
df["die_x"] = device_data.die.x
df["die_y"] = device_data.die.y
df["wafer_id"] = device_data.die.wafer.wafer_id
df["cell_id"] = device_data.device.cell.cell_id
df["parent_cell_id"] = device_data.device.parent_cell.cell_id
dfs_all.append(df)
dfs_all = pd.concat(dfs_all) # You can concatenate all dataFrames together
dfs_all
| wavelength | output_power | polyfit | device_id | die_x | die_y | wafer_id | cell_id | parent_cell_id | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1500.000000 | 0.031264 | 6.0 | RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_2... | 0 | 0 | 6d4c615ff105 | cutback_rib_assembled_MFalse_W0p3_L0 | RibLoss |
| 1 | 1500.833333 | 0.031813 | 6.0 | RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_2... | 0 | 0 | 6d4c615ff105 | cutback_rib_assembled_MFalse_W0p3_L0 | RibLoss |
| 2 | 1501.666667 | 0.036398 | 6.0 | RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_2... | 0 | 0 | 6d4c615ff105 | cutback_rib_assembled_MFalse_W0p3_L0 | RibLoss |
| 3 | 1502.500000 | 0.044324 | 6.0 | RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_2... | 0 | 0 | 6d4c615ff105 | cutback_rib_assembled_MFalse_W0p3_L0 | RibLoss |
| 4 | 1503.333333 | 0.039984 | 6.0 | RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_2... | 0 | 0 | 6d4c615ff105 | cutback_rib_assembled_MFalse_W0p3_L0 | RibLoss |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 116 | 1596.666667 | 0.041625 | 6.0 | RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_2... | 0 | 0 | 6d4c615ff105 | cutback_rib_assembled_MFalse_W0p3_L0 | RibLoss |
| 117 | 1597.500000 | 0.043175 | 6.0 | RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_2... | 0 | 0 | 6d4c615ff105 | cutback_rib_assembled_MFalse_W0p3_L0 | RibLoss |
| 118 | 1598.333333 | 0.036865 | 6.0 | RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_2... | 0 | 0 | 6d4c615ff105 | cutback_rib_assembled_MFalse_W0p3_L0 | RibLoss |
| 119 | 1599.166667 | 0.034557 | 6.0 | RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_2... | 0 | 0 | 6d4c615ff105 | cutback_rib_assembled_MFalse_W0p3_L0 | RibLoss |
| 120 | 1600.000000 | 0.032933 | 6.0 | RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_2... | 0 | 0 | 6d4c615ff105 | cutback_rib_assembled_MFalse_W0p3_L0 | RibLoss |
121 rows × 9 columns
Advanced queries#
To build advanced queries to filter metadata you can use the attribute_filter method. You can also use or_, and_ for conditional clauses.
Conditional filter#
data_tuples = dd.get_data_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.Device.device_id
== "RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_20150_60150",
]
)
len(data_tuples)
21
By default anything you pass to the list joins the clauses as and_
data_tuples = dd.get_data_by_query(
[
dd.and_(
dd.Project.project_id == PROJECT_ID,
dd.Device.device_id
== "RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_20150_60150",
)
]
)
len(data_tuples)
21
You can also use an OR condition.
The query below filters by project name PROJECT_ID AND either device name (rib_0p3 OR rib_0p5).
Because there are 21 measurements per device, one for each die, if you look for two specific devices, you will get 42 measurements.
data_tuples = dd.get_data_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.or_(
dd.Device.device_id
== "RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_20150_60150",
dd.Device.device_id
== "RibLoss_cutback_rib_assembled_MFalse_W0p5_L0_20250_492247",
),
]
)
len(data_tuples)
21
You can also combine conditionals.
In the example below, you can get all the data for the specified project name, device name and either die coordinate.
data_tuples = dd.get_data_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.Device.device_id
== "RibLoss_cutback_rib_assembled_MFalse_W0p3_L0_20150_60150",
dd.or_(
dd.and_(dd.Die.x == 1, dd.Die.y == 1), # either die 1,1
dd.and_(dd.Die.x == 0, dd.Die.y == 0), # or die 0,0
),
]
)
len(data_tuples)
2
Attribute filter#
You can filter attributes of any data models by passing a list of conditions as shown below.
You can use dd.Cell/dd.Wafer/..., key, value
You can only filter for values that are str, bool, int, float.
The query below filters Cells with waveguide width 0.3um that are in a particular project and die coordinates.
device_data_objects = dd.get_data_by_query(
[
dd.attribute_filter(
dd.Cell, "width_um", 0.3
), # checking for values in JSON attributes
dd.Project.project_id == PROJECT_ID,
dd.Die.x == 0,
dd.Die.y == 0,
]
)
len(device_data_objects)
6
You can combine conditional and attribute filter clauses together.
You can get the data for Cells for a given project with length_um 0um and either width_um 0.3um or 0.5um
data_tuples = dd.get_data_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.attribute_filter(dd.Cell, "length_um", 0),
dd.or_(
dd.attribute_filter(dd.Cell, "width_um", 0.3),
dd.attribute_filter(dd.Cell, "width_um", 0.5),
),
]
)
len(data_tuples)
42
