Upload Measurements and run analysis#
Now you will post your measurement data and analysis to the database via the DoData app.
If you are running the tutorials in DoLab, the following instructions are not necessary and you can skip directly to the next cell.
You will need to authenticate to the database with your username and password. To make this easy, you can create a file called .env in this folder and complete it with your organization’s URL and authentication information as follows:
dodata_url=https://animal.doplaydo.com
dodata_user=demo
dodata_password=yours
dodata_db=animal.dodata.db.doplaydo.com
dodata_db_user=full_access
dodata_db_password=yours
If you haven’t defined a .env file or saved your credentials to your environment variables, you will be prompted for your credentials now.
import doplaydo.dodata as dd
import pandas as pd
from pathlib import Path
from tqdm.auto import tqdm
import requests
import getpass
from httpx import HTTPStatusError
username = getpass.getuser()
Let’s now create a project.
In normal circumstances, everyone will be sharing and contributing to a project. In this demo, however, we want to keep your project separate from other users for clarity, so we will append your username to the project id. This way you can also safely delete and recreate projects without creating issues for others. If you prefer though, you can change the PROJECT_ID to anything you like. Just be sure to update it in the subsequent notebooks of this tutorial as well.
PROJECT_ID = f"rings-{username}"
MEASUREMENTS_PATH = Path("6d4c615ff105/")
If you’ve been running this demo multiple times, you might already have one with the same name. Let’s delete it so you can start fresh.
try:
dd.project.delete(project_id=PROJECT_ID).text
except HTTPStatusError:
pass
New project#
You can create the project, upload the design manifest, and upload the wafer definitions through the Webapp as well as programmatically using this notebook
Upload Project#
You can create a new project and extract all cells & devices below for the RidgeLoss and RibLoss
The expressions are regex expressions. For intro and testing your regexes you can check out regex101
To only extract top cells set max_hierarchy_lvl=-1 and min_hierarchy_lvl=-1
To disable extraction use a max_hierarchy_lvl < min_hierarchy_lvl
Whitelists take precedence over blacklists, so if you define both, it uses only the whitelist.
cell_extraction = [
dd.project.Extraction(
cell_id="rings",
cell_white_list=["^RingDouble"],
min_hierarchy_lvl=0,
max_hierarchy_lvl=0,
)
]
dd.project.create(
project_id=PROJECT_ID,
eda_file="test_chip.gds",
lyp_file="generic.lyp",
cell_extractions=cell_extraction,
).text
'{"success":20}'
Upload Design Manifest#
The design manifest is a CSV file that includes all the cell names, the cell settings, a list of analysis to trigger, and a list of settings for each analysis.
dm = pd.read_csv("design_manifest.csv")
dm
| cell | x | y | radius_um | gap_um | analysis | analysis_parameters | |
|---|---|---|---|---|---|---|---|
| 0 | RingDouble-20-0.25- | 331580 | 121311 | 20 | 0.25 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 1 | RingDouble-20-0.2- | 331580 | 285371 | 20 | 0.20 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 2 | RingDouble-20-0.15- | 331580 | 449331 | 20 | 0.15 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 3 | RingDouble-10-0.2- | 331480 | 613191 | 10 | 0.20 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 4 | RingDouble-10-0.15- | 331480 | 757151 | 10 | 0.15 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 5 | RingDouble-10-0.1- | 331480 | 901011 | 10 | 0.10 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 6 | RingDouble-5-0.2- | 331480 | 1044771 | 5 | 0.20 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 7 | RingDouble-5-0.15- | 331480 | 1178731 | 5 | 0.15 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 8 | RingDouble-5-0.1- | 331480 | 1312591 | 5 | 0.10 | [fsr] | [{"height": -0.01, "distance": 20}] |
As you don’t have an analysis defined yet you need to drop the columns that allow you to run automated analysis on measurement upload.
dm = dm.drop(columns=["analysis", "analysis_parameters"])
dm
| cell | x | y | radius_um | gap_um | |
|---|---|---|---|---|---|
| 0 | RingDouble-20-0.25- | 331580 | 121311 | 20 | 0.25 |
| 1 | RingDouble-20-0.2- | 331580 | 285371 | 20 | 0.20 |
| 2 | RingDouble-20-0.15- | 331580 | 449331 | 20 | 0.15 |
| 3 | RingDouble-10-0.2- | 331480 | 613191 | 10 | 0.20 |
| 4 | RingDouble-10-0.15- | 331480 | 757151 | 10 | 0.15 |
| 5 | RingDouble-10-0.1- | 331480 | 901011 | 10 | 0.10 |
| 6 | RingDouble-5-0.2- | 331480 | 1044771 | 5 | 0.20 |
| 7 | RingDouble-5-0.15- | 331480 | 1178731 | 5 | 0.15 |
| 8 | RingDouble-5-0.1- | 331480 | 1312591 | 5 | 0.10 |
dm.to_csv("design_manifest_without_analysis.csv", index=False)
dd.project.upload_design_manifest(
project_id=PROJECT_ID, filepath="design_manifest_without_analysis.csv"
).text
'{"success":200}'
dd.project.download_design_manifest(
project_id=PROJECT_ID, filepath="design_manifest_downloaded.csv"
)
PosixPath('design_manifest_downloaded.csv')
Upload Wafer Definitions#
The wafer definition is a JSON file where you can define the wafer names and die names and location for each wafer.
dd.project.upload_wafer_definitions(
project_id=PROJECT_ID, filepath="wafer_definitions.json"
).text
'{"success":200}'
Upload data#
Your Tester can output the data in JSON files. It does not need to be Python.
You can get all paths which have measurement data within the test path.
data_files = list(MEASUREMENTS_PATH.glob("**/data.json"))
print(data_files[0].parts)
('6d4c615ff105', '6_6', 'rings_RingDouble-10-0.15-_331480_757151', 'data.json')
You should define a plotting per measurement type in python. Your plots can evolve over time even for the same measurement type.
Required:
- x_name (str): x-axis name
- y_name (str): y-axis name
- x_col (str): x-column to plot
- y_col (list[str]): y-column(s) to plot; can be multiple
Optional:
- scatter (bool): whether to plot as scatter as opposed to line traces
- x_units (str): x-axis units
- y_units (str): y-axis units
- x_log_axis (bool): whether to plot the x-axis on log scale
- y_log_axis (bool): whether to plot the y-axis on log scale
- x_limits (list[int, int]): clip x-axis data using these limits as bounds (example: [10, 100])
- y_limits (list[int, int]): clip y-axis data using these limits as bounds (example: [20, 100])
- sort_by (dict[str, bool]): columns to sort data before plotting. Boolean specifies whether to sort each column in ascending order.
(example: {"wavelegths": True, "optical_power": False})
- grouping (dict[str, int]): columns to group data before plotting. Integer specifies decimal places to round each column.
Different series will be plotted for unique combinations of x column, y column(s), and rounded column values.
(example: {"port": 1, "attenuation": 2})
spectrum_measurement_type = dd.api.device_data.PlottingKwargs(
x_name="wavelength",
y_name="output_power",
x_col="wavelength",
y_col=["output_power"],
)
Upload measurements#
You can now upload measurement data.
This is a bare bones example, in a production setting, you can also add validation, logging, and error handling to ensure a smooth operation.
Every measurement you upload will trigger all the analysis that you defined in the design manifest.
NUMBER_OF_THREADS = 1 if "127" in dd.settings.dodata_url else dd.settings.n_threads
wafer_set = set()
die_set = set()
NUMBER_OF_THREADS
1
if NUMBER_OF_THREADS == 1:
for path in tqdm(data_files):
wafer_id = path.parts[0]
die_x, die_y = path.parts[1].split("_")
r = dd.api.device_data.upload(
file=path,
project_id=PROJECT_ID,
wafer_id=wafer_id,
die_x=die_x,
die_y=die_y,
device_id=path.parts[2],
data_type="measurement",
plotting_kwargs=spectrum_measurement_type,
)
wafer_set.add(wafer_id)
die_set.add(path.parts[2])
r.raise_for_status()
data_files = list(MEASUREMENTS_PATH.glob("**/data.json"))
project_ids = []
device_ids = []
die_ids = []
die_xs = []
die_ys = []
wafer_ids = []
plotting_kwargs = []
data_types = []
for path in data_files:
device_id = path.parts[2]
die_id = path.parts[1]
die_x, die_y = die_id.split("_")
wafer_id = path.parts[0]
device_ids.append(device_id)
die_ids.append(die_id)
die_xs.append(die_x)
die_ys.append(die_y)
die_set.add(die_id)
wafer_ids.append(wafer_id)
plotting_kwargs.append(spectrum_measurement_type)
project_ids.append(PROJECT_ID)
data_types.append("measurement")
wafer_set.add(wafer_id)
if NUMBER_OF_THREADS > 1:
dd.device_data.upload_multi(
files=data_files,
project_ids=project_ids,
wafer_ids=wafer_ids,
die_xs=die_xs,
die_ys=die_ys,
device_ids=device_ids,
data_types=data_types,
plotting_kwargs=plotting_kwargs,
progress_bar=True,
)
Analysis#
You can run analysis at 3 different levels. For example to extract:
Device: averaged power envelope over certain number of samples.
Die: fit the propagation loss as a function of length.
Wafer: Define the Upper and Lower Spec limits for Known Good Die (KGD)

To upload custom analysis functions to the DoData server, follow these simplified guidelines:
Input:
Begin with a unique identifier (device_data_id, die_id, wafer_id) as the first argument.
Add necessary keyword arguments for the analysis.
Output: Dictionary
output: Return a simple, one-level dictionary. All values must be serializable. Avoid using numpy or pandas; convert to lists if needed.
summary_plot: Provide a summary plot, either as a matplotlib figure or io.BytesIO object.
attributes: Add a serializable dictionary of the analysis settings.
device_data_id/die_id/wafer_id: Include the used identifier (device_data_id, die_id, wafer_id).
Device analysis#
You can either trigger analysis automatically by defining it in the design manifest, using the UI or using the Python DoData library.
from IPython.display import Code, display, Image
import doplaydo.dodata as dd
display(Code(dd.config.Path.analysis_functions_device_fsr))
"""Free spectral range (FSR) analysis."""
from typing import Any
import numpy as np
from matplotlib import pyplot as plt
from scipy.signal import find_peaks
import doplaydo.dodata as dd
def find_resonance_peaks(
y, height: float = 0.1, threshold: None | float = None, distance: float | None = 10
):
"""Find the resonance peaks in the ring resonator response.
'height' and 'distance' can be adjusted based on the specifics of your data.
Args:
y: ndarray
height : number or ndarray or sequence, optional
Required height of peaks. Either a number, ``None``, an array matching
`x` or a 2-element sequence of the former. The first element is
always interpreted as the minimal and the second, if supplied, as the
maximal required height.
threshold : number or ndarray or sequence, optional
Required threshold of peaks, the vertical distance to its neighboring
samples. Either a number, ``None``, an array matching `x` or a
2-element sequence of the former. The first element is always
interpreted as the minimal and the second, if supplied, as the maximal
required threshold.
distance : number, optional
Required minimal horizontal distance (>= 1) in samples between
neighbouring peaks. Smaller peaks are removed first until the condition
is fulfilled for all remaining peaks.
"""
if height < 0:
y = -y
height = abs(height)
peaks, _ = find_peaks(y, height=height, distance=distance)
return peaks
def remove_baseline(wavelength: np.ndarray, power: np.ndarray, deg: int = 4):
"""Return power corrected without baseline.
Fit a polynomial ``p(x) = p[0] * x**deg + ... + p[deg]`` of degree `deg`
"""
pfit = np.polyfit(wavelength - np.mean(wavelength), power, deg)
power_baseline = np.polyval(pfit, wavelength - np.mean(wavelength))
power_corrected = power - power_baseline
power_corrected = power_corrected + max(power_baseline) - max(power)
return power_corrected
def run(
device_data_pkey: int,
height: float = -0.1,
threshold: None | float = None,
distance: float | None = 10,
xkey: str = "wavelength",
ykey: str = "output_power",
) -> dict[str, Any]:
"""Returns the Free spectral range (FSR) for a resonator.
Args:
device_data_pkey: The pkey of the device data to be analyzed.
height: Required height of peaks. <0 for minima.
threshold: Required threshold of peaks, the vertical distance to its neighboring.
distance: Required minimal horizontal distance (>= 1) in samples between.
xkey: The key of the x data in the device data.
ykey: The key of the y data in the device data.
"""
data = dd.get_data_by_pkey(device_data_pkey)
if data is None:
raise ValueError(f"Device data with pkey {device_data_pkey} not found.")
if xkey not in data:
raise ValueError(
f"Device data with pkey {device_data_pkey} does not have xkey {xkey!r}."
)
if ykey not in data:
raise ValueError(
f"Device data with pkey {device_data_pkey} does not have ykey {ykey!r}."
)
x = data[xkey].values
y = data[ykey].values
spectrum = y
spectrum_normalized = remove_baseline(wavelength=x, power=y, deg=4)
peaks = find_resonance_peaks(
spectrum_normalized, height=height, distance=distance, threshold=threshold
)
if not peaks.any():
raise ValueError(
f"No peaks found for device data with pkey {device_data_pkey}, adjust height {height}."
)
peak_frequencies = x[peaks]
fsr = np.diff(peak_frequencies)
fsr_mean = np.mean(fsr)
fsr_std = np.std(fsr)
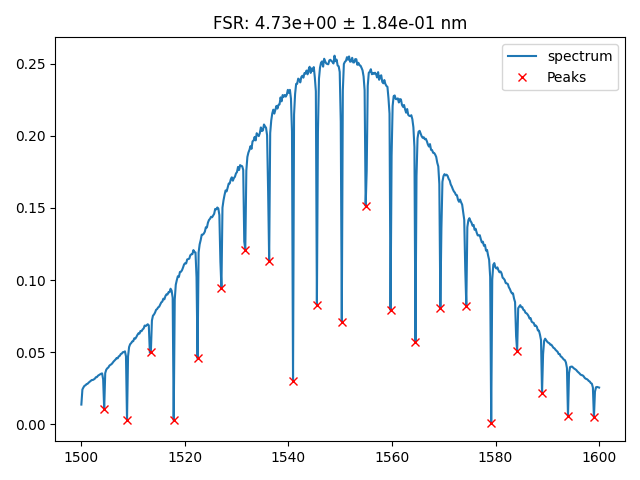
fig = plt.figure()
plt.plot(x, spectrum, label="spectrum")
plt.plot(x[peaks], spectrum[peaks], "x", color="red", label="Peaks")
plt.legend()
plt.title(f"FSR: {fsr_mean:.2e} ± {fsr_std:.2e} nm")
if len(peaks) < 2:
raise ValueError(
f"Only one peak found for device data with pkey {device_data_pkey}, adjust height {height}."
)
if len(peaks) > 200:
raise ValueError(
f"More than 200 peaks found for device data with pkey {device_data_pkey}, adjust height {height}."
)
return dict(
output={
"fsr_mean": float(fsr_mean),
"fsr_std": float(fsr_std),
"peaks": x[peaks].tolist() or None,
"fsr": fsr.tolist() or None,
},
summary_plot=fig,
device_data_pkey=device_data_pkey,
)
if __name__ == "__main__":
d = run(82766, height=-0.01)
print(d["output"]["fsr_mean"])
You can easily get a device pkey to try your device analsys:
device_data = dd.get_data_objects_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.attribute_filter(dd.Cell, "radius_um", 20),
dd.attribute_filter(dd.Cell, "gap_um", 0.2),
],
limit=1,
)[0]
device_data.pkey
6427
device_data.device.device_id
'rings_RingDouble-20-0.2-_331580_285371'
response = dd.api.analysis_functions.validate(
analysis_function_id="device_fsr",
function_path=dd.config.Path.analysis_functions_device_fsr,
test_model_pkey=device_data.pkey,
target_model_name="device_data",
parameters=dict(height=-0.02),
)
Image(response.content)
Headers({'server': 'nginx/1.22.1', 'date': 'Fri, 28 Feb 2025 21:02:34 GMT', 'content-type': 'image/png', 'transfer-encoding': 'chunked', 'connection': 'keep-alive', '_output': '{"fsr_mean": 4.729458917835666, "fsr_std": 0.18367036853529464, "peaks": [1504.4088176352707, 1508.8176352705411, 1513.426853707415, 1517.8356713426856, 1522.4448897795592, 1527.0541082164327, 1531.6633266533065, 1536.2725450901803, 1540.881763527054, 1545.4909819639279, 1550.3006012024048, 1554.9098196392786, 1559.7194388777555, 1564.5290581162326, 1569.3386773547095, 1574.3486973947895, 1579.1583166332666, 1584.1683366733469, 1588.9779559118238, 1593.987975951904, 1598.997995991984], "fsr": [4.408817635270452, 4.609218436873789, 4.40881763527068, 4.609218436873562, 4.609218436873562, 4.609218436873789, 4.609218436873789, 4.609218436873789, 4.609218436873789, 4.809619238476898, 4.609218436873789, 4.809619238476898, 4.809619238477126, 4.809619238476898, 5.010020040080008, 4.809619238477126, 5.010020040080235, 4.809619238476898, 5.010020040080235, 5.010020040080008]}', '_attributes': '{}', '_device_data_pkey': '6427', 'strict-transport-security': 'max-age=63072000'})
response = dd.api.analysis_functions.validate_and_upload(
analysis_function_id="device_fsr",
function_path=dd.config.Path.analysis_functions_device_fsr,
test_model_pkey=device_data.pkey,
target_model_name="device_data",
parameters=dict(height=-0.02),
)
Headers({'server': 'nginx/1.22.1', 'date': 'Fri, 28 Feb 2025 21:02:35 GMT', 'content-type': 'image/png', 'transfer-encoding': 'chunked', 'connection': 'keep-alive', '_output': '{"fsr_mean": 4.729458917835666, "fsr_std": 0.18367036853529464, "peaks": [1504.4088176352707, 1508.8176352705411, 1513.426853707415, 1517.8356713426856, 1522.4448897795592, 1527.0541082164327, 1531.6633266533065, 1536.2725450901803, 1540.881763527054, 1545.4909819639279, 1550.3006012024048, 1554.9098196392786, 1559.7194388777555, 1564.5290581162326, 1569.3386773547095, 1574.3486973947895, 1579.1583166332666, 1584.1683366733469, 1588.9779559118238, 1593.987975951904, 1598.997995991984], "fsr": [4.408817635270452, 4.609218436873789, 4.40881763527068, 4.609218436873562, 4.609218436873562, 4.609218436873789, 4.609218436873789, 4.609218436873789, 4.609218436873789, 4.809619238476898, 4.609218436873789, 4.809619238476898, 4.809619238477126, 4.809619238476898, 5.010020040080008, 4.809619238477126, 5.010020040080235, 4.809619238476898, 5.010020040080235, 5.010020040080008]}', '_attributes': '{}', '_device_data_pkey': '6427', 'strict-transport-security': 'max-age=63072000'})
Trigger all analyses for all device data#
You have 64 dies and you will get 64 analysis, one for each device.
device_data_objects = dd.get_data_objects_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.attribute_filter(dd.Cell, "gap_um", 0.2),
]
)
device_data_pkeys = [d.pkey for d in device_data_objects]
params = [dict(height=-0.02)] * len(device_data_objects)
analyses = dd.api.analysis.trigger_device_data_multi(
device_data_pkeys=device_data_pkeys,
analysis_function_id="device_fsr",
n_threads=2,
parameters=params,
progress_bar=True,
)
len(analyses)
192
device_data_objects[0].device.device_id
'rings_RingDouble-5-0.2-_331480_1044771'
len(set(device_data_pkeys))
192
Die analysis#
You can aggregate any metric for a die analysis, for example, we had 3 different ring radius. Each of which will have a different FSR.
device_attributes = {"radius_um": 20, "gap_um": 0.2}
response = dd.api.analysis_functions.validate(
analysis_function_id="die_aggregate",
function_path=dd.config.Path.analysis_functions_die_aggregate,
test_model_pkey=device_data_objects[0].die.pkey,
target_model_name="die",
parameters=dict(
device_attributes=device_attributes,
device_analysis_function="device_fsr",
metric="fsr_mean",
),
)
Image(response.content)
Headers({'server': 'nginx/1.22.1', 'date': 'Fri, 28 Feb 2025 21:03:00 GMT', 'content-type': 'image/png', 'transfer-encoding': 'chunked', 'connection': 'keep-alive', '_output': '{"mean": 4.729458917835666, "std": 0.0, "median": 4.729458917835666}', '_attributes': '{}', '_die_pkey': '766', 'strict-transport-security': 'max-age=63072000'})
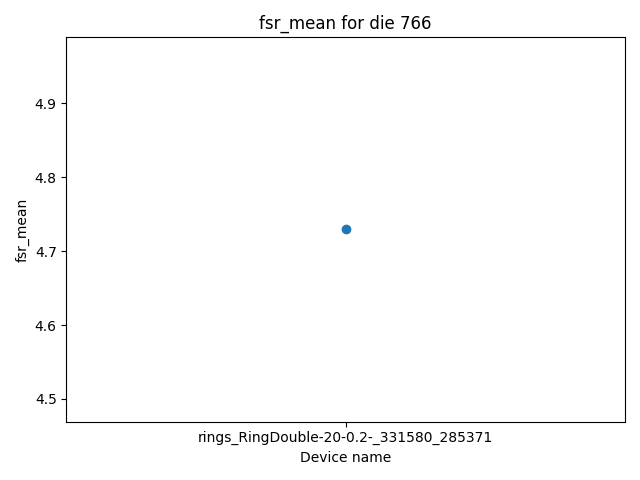
device_data_objects[0].die.pkey
766
response = dd.api.analysis_functions.validate_and_upload(
analysis_function_id="die_aggregate",
function_path=dd.config.Path.analysis_functions_die_aggregate,
test_model_pkey=device_data_objects[0].die.pkey,
target_model_name="die",
parameters=dict(
device_attributes=device_attributes,
device_analysis_function="device_fsr",
metric="fsr_mean",
),
)
Headers({'server': 'nginx/1.22.1', 'date': 'Fri, 28 Feb 2025 21:03:01 GMT', 'content-type': 'image/png', 'transfer-encoding': 'chunked', 'connection': 'keep-alive', '_output': '{"mean": 4.729458917835666, "std": 0.0, "median": 4.729458917835666}', '_attributes': '{}', '_die_pkey': '766', 'strict-transport-security': 'max-age=63072000'})
device_data_objects[0].device.device_id
'rings_RingDouble-5-0.2-_331480_1044771'
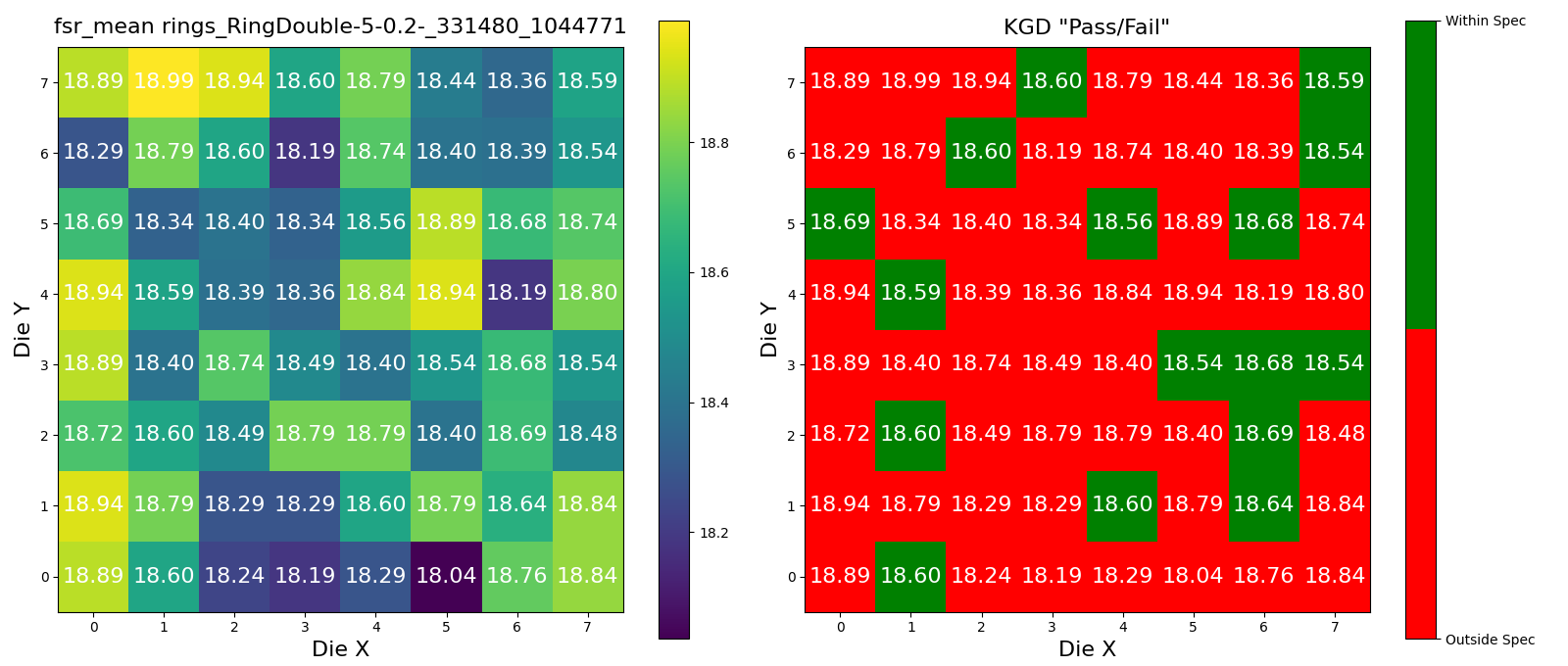
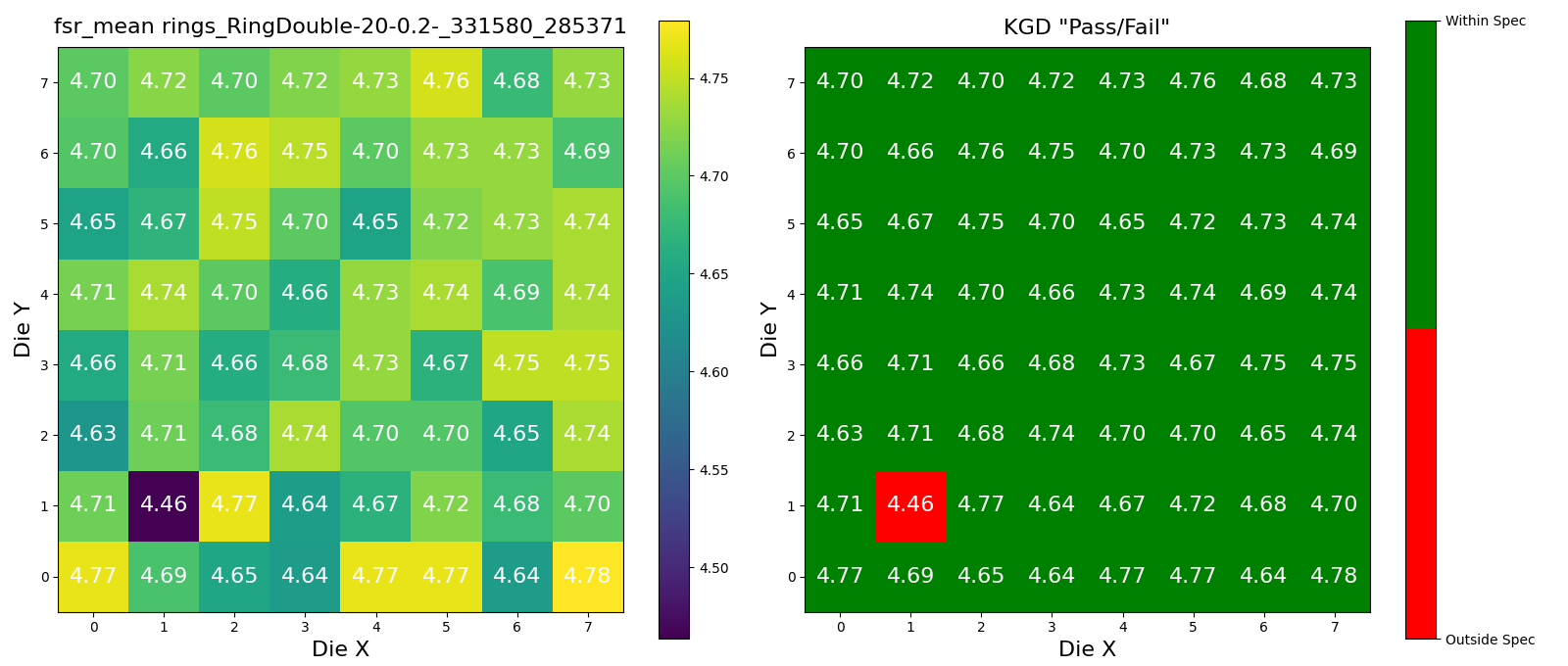
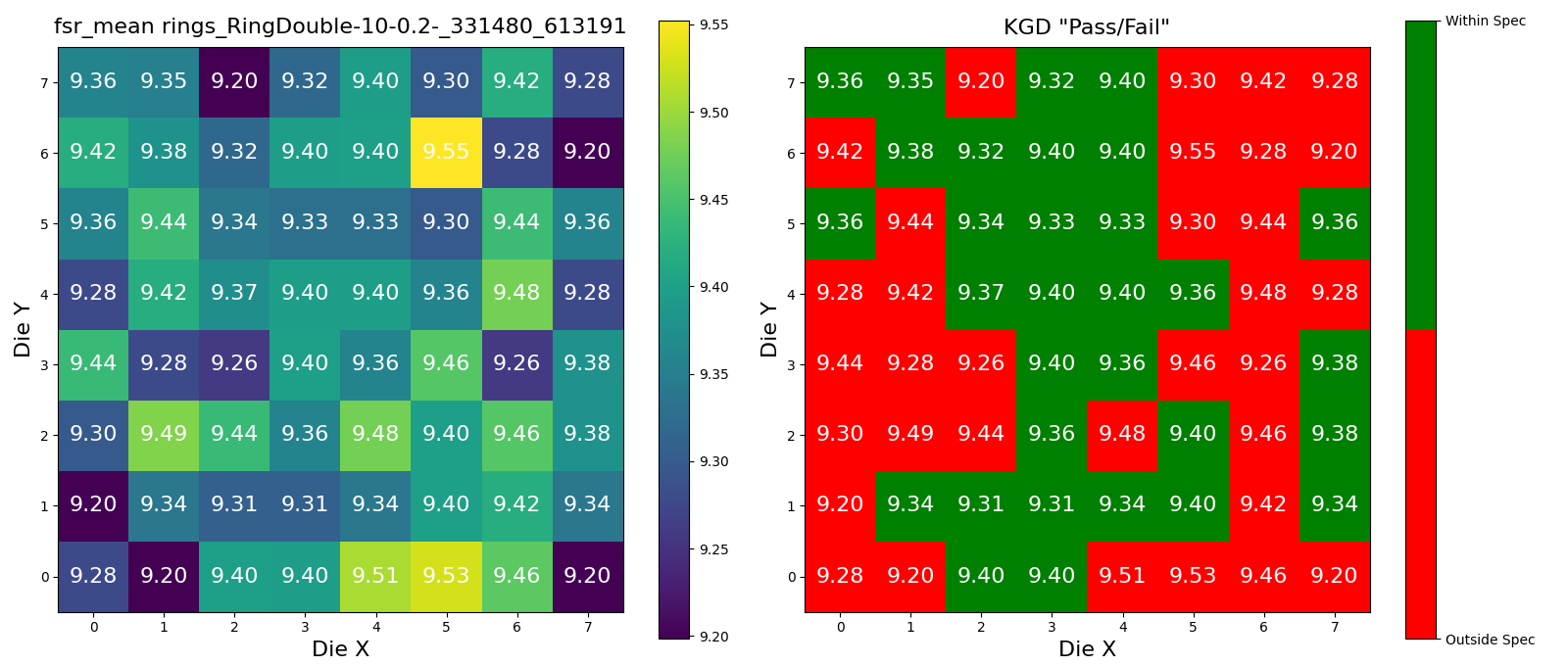
Wafer Analysis#
Lets Define the Upper and Lower Spec limits for Known Good Die (KGD).
Lets find a wafer pkey for this project, so that we can trigger the die analysis on it.
wafer_pkey = device_data_objects[0].die.wafer.pkey
wafer_id = device_data_objects[0].die.wafer.wafer_id
wafer_id
'6d4c615ff105'
parameters = dict(
device_id=device_data_objects[0].device.device_id,
lower_spec=4.60,
upper_spec=4.75,
analysis_function_id="device_fsr",
metric="fsr_mean",
)
response = dd.api.analysis_functions.validate(
analysis_function_id="wafer_aggregate",
function_path=dd.config.Path.analysis_functions_wafer_device_data_id,
test_model_pkey=wafer_pkey,
target_model_name="wafer",
parameters=parameters,
)
Image(response.content)
Headers({'server': 'nginx/1.22.1', 'date': 'Fri, 28 Feb 2025 21:03:01 GMT', 'content-type': 'image/png', 'transfer-encoding': 'chunked', 'connection': 'keep-alive', '_output': '{"result": [18.386773547094208, 18.557114228456932, 18.787575150300654, 18.39679358717435, 18.48697394789582, 18.5971943887776, 18.687374749499043, 18.63727454909821, 18.39679358717435, 18.937875751503043, 18.537074148296597, 18.75751503006013, 18.837675350701375, 18.677354709418843, 18.73747494989982, 18.39679358717435, 18.937875751502986, 18.787575150300597, 18.33667334669343, 18.787575150300597, 18.537074148296654, 18.5971943887776, 18.937875751502986, 18.587174348697374, 18.18637274549104, 18.88777555110221, 18.48697394789582, 18.787575150300597, 18.597194388777552, 18.286573146292653, 18.47695390781564, 18.35671342685373, 18.717434869739463, 18.73747494989982, 18.88777555110221, 18.687374749498986, 18.797595190380754, 18.88777555110221, 18.39679358717435, 18.587174348697374, 18.537074148296597, 18.787575150300597, 18.39679358717435, 18.286573146292596, 18.98797595190382, 18.356713426853684, 18.28657314629254, 18.336673346693374, 18.83767535070143, 18.18637274549104, 18.677354709418843, 18.38677354709415, 18.837675350701375, 18.5971943887776, 18.186372745490928, 18.73747494989982, 18.436873747494975, 18.937875751502986, 18.036072144288575, 18.88777555110221, 18.597194388777552, 18.787575150300597, 18.236472945891762, 18.286573146292653]}', '_attributes': '{}', '_wafer_pkey': '25', 'strict-transport-security': 'max-age=63072000'})
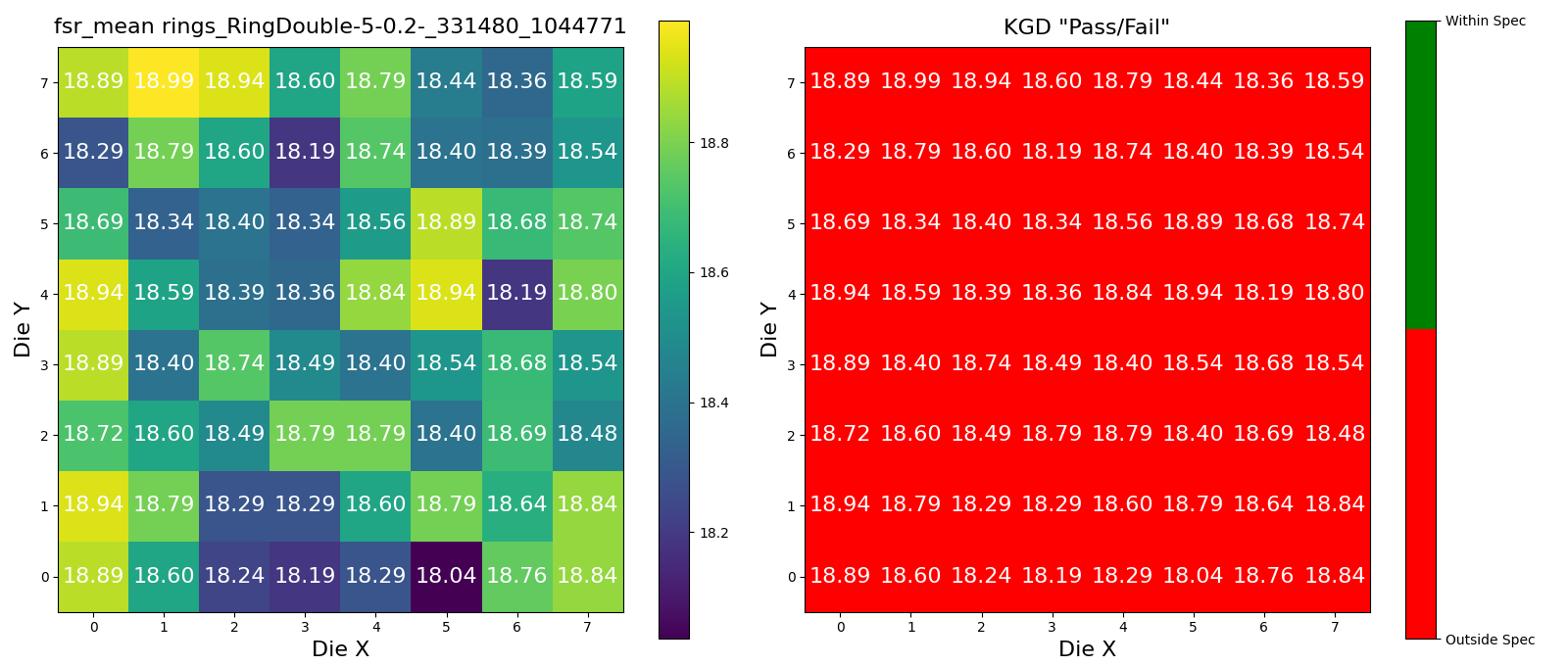
response = dd.api.analysis_functions.validate_and_upload(
analysis_function_id="wafer_aggregate",
function_path=dd.config.Path.analysis_functions_wafer_device_data_id,
test_model_pkey=wafer_pkey,
target_model_name="wafer",
parameters=parameters,
)
Headers({'server': 'nginx/1.22.1', 'date': 'Fri, 28 Feb 2025 21:03:02 GMT', 'content-type': 'image/png', 'transfer-encoding': 'chunked', 'connection': 'keep-alive', '_output': '{"result": [18.386773547094208, 18.557114228456932, 18.787575150300654, 18.39679358717435, 18.48697394789582, 18.5971943887776, 18.687374749499043, 18.63727454909821, 18.39679358717435, 18.937875751503043, 18.537074148296597, 18.75751503006013, 18.837675350701375, 18.677354709418843, 18.73747494989982, 18.39679358717435, 18.937875751502986, 18.787575150300597, 18.33667334669343, 18.787575150300597, 18.537074148296654, 18.5971943887776, 18.937875751502986, 18.587174348697374, 18.18637274549104, 18.88777555110221, 18.48697394789582, 18.787575150300597, 18.597194388777552, 18.286573146292653, 18.47695390781564, 18.35671342685373, 18.717434869739463, 18.73747494989982, 18.88777555110221, 18.687374749498986, 18.797595190380754, 18.88777555110221, 18.39679358717435, 18.587174348697374, 18.537074148296597, 18.787575150300597, 18.39679358717435, 18.286573146292596, 18.98797595190382, 18.356713426853684, 18.28657314629254, 18.336673346693374, 18.83767535070143, 18.18637274549104, 18.677354709418843, 18.38677354709415, 18.837675350701375, 18.5971943887776, 18.186372745490928, 18.73747494989982, 18.436873747494975, 18.937875751502986, 18.036072144288575, 18.88777555110221, 18.597194388777552, 18.787575150300597, 18.236472945891762, 18.286573146292653]}', '_attributes': '{}', '_wafer_pkey': '25', 'strict-transport-security': 'max-age=63072000'})
Now that you have uploaded the wafer analysis function you can trigger the wafer analysis for all the wafers so you can store and visualize the wafermaps in the DoData website.
device_data, df = dd.get_data_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.attribute_filter(dd.Cell, "radius_um", 20),
dd.attribute_filter(dd.Cell, "gap_um", 0.2),
],
limit=1,
)[0]
device_data.device.device_id
'rings_RingDouble-20-0.2-_331580_285371'
device_data20, df = dd.get_data_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.attribute_filter(dd.Cell, "radius_um", 20),
dd.attribute_filter(dd.Cell, "gap_um", 0.2),
],
limit=1,
)[0]
device_data10, df = dd.get_data_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.attribute_filter(dd.Cell, "radius_um", 10),
dd.attribute_filter(dd.Cell, "gap_um", 0.2),
],
limit=1,
)[0]
device_data5, df = dd.get_data_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.attribute_filter(dd.Cell, "radius_um", 5),
dd.attribute_filter(dd.Cell, "gap_um", 0.2),
],
limit=1,
)[0]
You can trigger the device analysis for rings with 200nm gaps and 20, 10 and 5um.
64 dies and 3 devices will trigger 192 device analysis.
device_data_objects = dd.get_data_objects_by_query(
[
dd.Project.project_id == PROJECT_ID,
dd.or_(
dd.Device.device_id == device_data20.device.device_id,
dd.Device.device_id == device_data10.device.device_id,
dd.Device.device_id == device_data5.device.device_id,
),
],
limit=1,
)
device_data_pkeys = [d.pkey for d in device_data_objects]
params = [dict(height=-0.02)] * len(device_data_objects)
analyses = dd.api.analysis.trigger_device_data_multi(
device_data_pkeys=device_data_pkeys,
analysis_function_id="device_fsr",
parameters=params,
progress_bar=True,
)
len(analyses)
1
wafer_id = device_data5.die.wafer.wafer_id
wafer_set = set([wafer_id])
# 5um radius rings
device_id = device_data5.device.device_id
for wafer in tqdm(wafer_set):
r = dd.analysis.trigger_wafer(
project_id=PROJECT_ID,
wafer_id=wafer,
analysis_function_id="wafer_aggregate",
parameters=dict(
device_id=device_id,
lower_spec=18.5,
upper_spec=18.7,
analysis_function_id="device_fsr",
metric="fsr_mean",
),
)
if r.status_code != 200:
raise requests.HTTPError(r.text)
# 10um radius rings
device_id = device_data10.device.device_id
for wafer in tqdm(wafer_set):
r = dd.analysis.trigger_wafer(
project_id=PROJECT_ID,
wafer_id=wafer,
analysis_function_id="wafer_aggregate",
parameters=dict(
device_id=device_id,
lower_spec=9.30,
upper_spec=9.40,
analysis_function_id="device_fsr",
metric="fsr_mean",
),
)
if r.status_code != 200:
raise requests.HTTPError(r.text)
# 20um radius rings
device_id = device_data20.device.device_id
for wafer in tqdm(wafer_set):
r = dd.analysis.trigger_wafer(
project_id=PROJECT_ID,
wafer_id=wafer,
analysis_function_id="wafer_aggregate",
parameters=dict(
device_id=device_id,
lower_spec=4.6,
upper_spec=4.8,
analysis_function_id="device_fsr",
metric="fsr_mean",
),
)
if r.status_code != 200:
raise requests.HTTPError(r.text)
plots = dd.analysis.get_wafer_analysis_plots(
project_id=PROJECT_ID,
wafer_id=wafer_id,
target_model="wafer",
)
len(plots)
3
for plot in plots:
display(plot)