Measurements#
In this tutorial you will generate some sample (fake) measurement data so you can post it to your project.
You can create a new folder and populate it with JSON files containing the fake measurement data for the whole wafer.
import json
import pandas as pd
import numpy as np
from pathlib import Path
import matplotlib.pyplot as plt
df = pd.read_csv("design_manifest.csv")
df
| cell | x | y | radius_um | gap_um | analysis | analysis_parameters | |
|---|---|---|---|---|---|---|---|
| 0 | RingDouble-20-0.25- | 331580 | 121311 | 20 | 0.25 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 1 | RingDouble-20-0.2- | 331580 | 285371 | 20 | 0.20 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 2 | RingDouble-20-0.15- | 331580 | 449331 | 20 | 0.15 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 3 | RingDouble-10-0.2- | 331480 | 613191 | 10 | 0.20 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 4 | RingDouble-10-0.15- | 331480 | 757151 | 10 | 0.15 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 5 | RingDouble-10-0.1- | 331480 | 901011 | 10 | 0.10 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 6 | RingDouble-5-0.2- | 331480 | 1044771 | 5 | 0.20 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 7 | RingDouble-5-0.15- | 331480 | 1178731 | 5 | 0.15 | [fsr] | [{"height": -0.01, "distance": 20}] |
| 8 | RingDouble-5-0.1- | 331480 | 1312591 | 5 | 0.10 | [fsr] | [{"height": -0.01, "distance": 20}] |
def ring(
wl: np.ndarray,
wl0: float,
neff: float,
ng: float,
ring_length: float,
coupling: float,
loss: float,
) -> np.ndarray:
"""Returns Frequency Domain Response of an all pass filter.
Args:
wl: wavelength in um.
wl0: center wavelength at which neff and ng are defined.
neff: effective index.
ng: group index.
ring_length: in um.
loss: dB/um.
"""
transmission = 1 - coupling
neff_wl = (
neff + (wl0 - wl) * (ng - neff) / wl0
) # we expect a linear behavior with respect to wavelength
out = np.sqrt(transmission) - 10 ** (-loss * ring_length * 1e-6 / 20.0) * np.exp(
2j * np.pi * neff_wl * ring_length / wl
)
out /= 1 - np.sqrt(transmission) * 10 ** (
-loss * ring_length * 1e-6 / 20.0
) * np.exp(2j * np.pi * neff_wl * ring_length / wl)
return abs(out) ** 2
def gaussian_grating_coupler_response(
peak_power, center_wavelength, bandwidth_1dB, wavelength
):
"""Calculate the response of a Gaussian grating coupler.
Args:
peak_power: The peak power of the response.
center_wavelength: The center wavelength of the grating coupler.
bandwidth_1dB: The 1 dB bandwidth of the coupler.
wavelength: The wavelength at which the response is evaluated.
Returns:
The power of the grating coupler response at the given wavelength.
"""
# Convert 1 dB bandwidth to standard deviation (sigma)
sigma = bandwidth_1dB / (2 * np.sqrt(2 * np.log(10)))
# Gaussian response calculation
response = peak_power * np.exp(
-0.5 * ((wavelength - center_wavelength) / sigma) ** 2
)
return response
nm = 1e-3
# Parameters
peak_power = 1.0
center_wavelength = 1550 * nm # Center wavelength in micrometers
bandwidth_1dB = 100 * nm
# Wavelength range: 100 nm around the center wavelength, converted to micrometers
wavelength_range = np.linspace(center_wavelength - 0.05, center_wavelength + 0.05, 500)
# Calculate the response for each wavelength
grating_spectrum = np.array(
[
gaussian_grating_coupler_response(
peak_power, center_wavelength, bandwidth_1dB, wl
)
for wl in wavelength_range
]
)
ring_spectrum = ring(
wavelength_range,
wl0=1.55,
neff=2.4,
ng=4.0,
ring_length=2 * np.pi * 10,
coupling=0.1,
loss=30e2, # 30 dB/cm = 30000 dB/m
)
spectrum = ring_spectrum * grating_spectrum
# Plotting
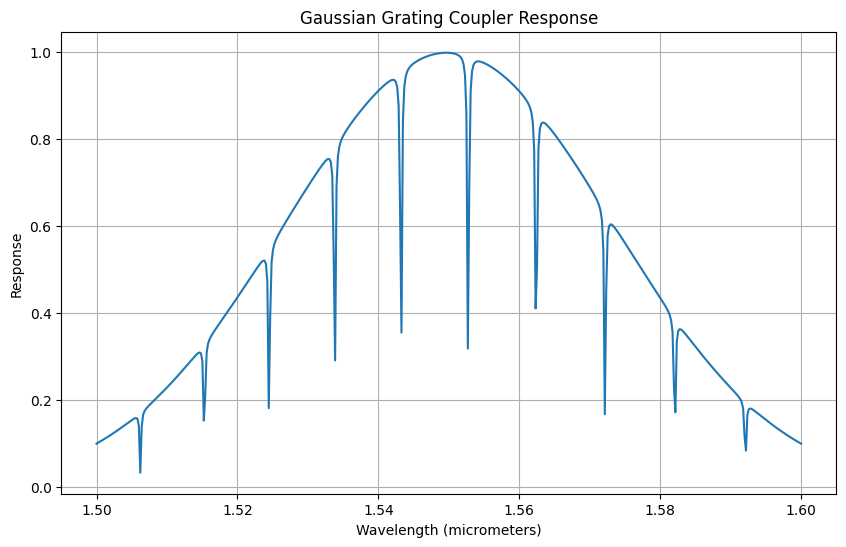
plt.figure(figsize=(10, 6))
plt.plot(wavelength_range, spectrum)
plt.title("Gaussian Grating Coupler Response")
plt.xlabel("Wavelength (micrometers)")
plt.ylabel("Response")
plt.grid(True)
plt.show()
for radius in [5, 10, 20]:
dB = 3 * (radius * 1e-4 * 2 * 3.14)
print(10 ** (-dB / 10))
0.9978333154992974
0.9956713255203204
0.9913613884633918
import numpy as np
from scipy.signal import find_peaks
def find_resonance_peaks(
y, height: float = 0.1, threshold: None | float = None, distance: float | None = 10
):
"""Find the resonance peaks in the ring resonator response.
'height' and 'distance' can be adjusted based on the specifics of your data.
Args:
y: ndarray
height : number or ndarray or sequence, optional
Required height of peaks. Either a number, ``None``, an array matching
`x` or a 2-element sequence of the former. The first element is
always interpreted as the minimal and the second, if supplied, as the
maximal required height.
threshold : number or ndarray or sequence, optional
Required threshold of peaks, the vertical distance to its neighboring
samples. Either a number, ``None``, an array matching `x` or a
2-element sequence of the former. The first element is always
interpreted as the minimal and the second, if supplied, as the maximal
required threshold.
distance : number, optional
Required minimal horizontal distance (>= 1) in samples between
neighbouring peaks. Smaller peaks are removed first until the condition
is fulfilled for all remaining peaks.
"""
if height < 0:
y = -y
height = abs(height)
peaks, _ = find_peaks(y, height=height, distance=distance)
return peaks
def calculate_FSR(x, peaks):
"""Calculate the Free Spectral Range (FSR) based on the found peaks."""
peak_frequencies = x[peaks]
fsr = np.diff(peak_frequencies)
return fsr
def remove_baseline(wavelength: np.ndarray, power: np.ndarray, deg: int = 4):
"""Return power corrected without baseline.
Fit a polynomial ``p(x) = p[0] * x**deg + ... + p[deg]`` of degree `deg`
"""
pfit = np.polyfit(wavelength - np.mean(wavelength), power, deg)
power_baseline = np.polyval(pfit, wavelength - np.mean(wavelength))
power_corrected = power - power_baseline
power_corrected = power_corrected + max(power_baseline) - max(power)
return power_corrected
x = wavelength_range
y = remove_baseline(wavelength=wavelength_range, power=spectrum, deg=4)
# Find resonance peaks
peaks = find_resonance_peaks(y, height=-0.01, distance=30, threshold=None)
y = spectrum
# Calculate FSR
fsr = calculate_FSR(x, peaks)
print("Resonance Peaks at:", x[peaks])
print("Free Spectral Range (FSR):", fsr)
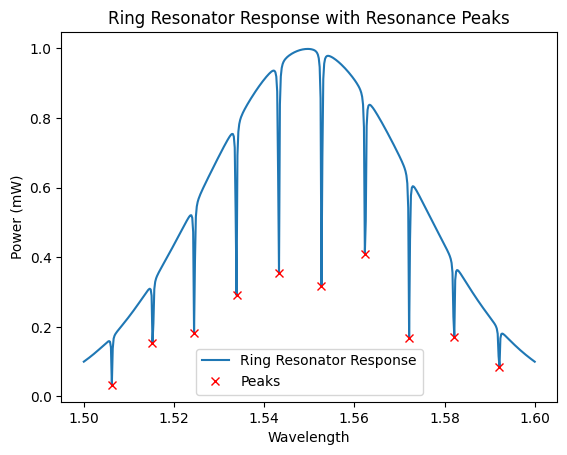
plt.plot(x, y, label="Ring Resonator Response")
plt.plot(x[peaks], y[peaks], "x", color="red", label="Peaks")
plt.title("Ring Resonator Response with Resonance Peaks")
plt.xlabel("Wavelength")
plt.ylabel("Power (mW)")
plt.legend()
plt.show()
Resonance Peaks at: [1.50621242 1.51523046 1.5244489 1.53386774 1.54328657 1.55270541
1.56232465 1.57214429 1.58216433 1.59218437]
Free Spectral Range (FSR): [0.00901804 0.00921844 0.00941884 0.00941884 0.00941884 0.00961924
0.00981964 0.01002004 0.01002004]
def compute_group_index(wavelength, length, fsr):
"""Compute the group index (ng) of a ring resonator.
Args:
wavelength: Central wavelength of the resonator (in meters).
length: Physical length of the ring resonator (in meters).
fsr: Free Spectral Range (in meters).
"""
ng = (wavelength**2) / (length * fsr)
return ng
compute_group_index(x[peaks][:-1], 2 * np.pi * 10, fsr)
array([4.00387596, 3.96387755, 3.92688841, 3.97556304, 4.02453747,
3.98894063, 3.95609914, 3.92586605, 3.97606843])
peaks.tolist()
[31, 76, 122, 169, 216, 263, 311, 360, 410, 460]
Generate wafer definitions#
You can define different wafer maps for each wafer.

wafer_definitions = Path("wafer_definitions.json")
wafers = ["6d4c615ff105"]
wafer_attributes = dict(
lot_id="lot1",
attributes=dict(process_type="low_loss", wafer_diameter=200, wafer_thickness=0.7),
)
dies = [{"x": x, "y": y, "id": x * 8 + y} for y in range(0, 8) for x in range(0, 8)]
# Wrap in a list with the wafer information
data = [
{"wafer": wafer_pkey, "dies": dies, **wafer_attributes} for wafer_pkey in wafers
]
with open(wafer_definitions, "w") as f:
json.dump(data, f, indent=2)
Generate and write spectrums#
You can easily generate some spectrum data and add some noise to make it look like a real measurement.
cwd = Path(".")
metadata = {"measurement_type": "Spectral MEAS", "temperature": 25}
noise_peak_to_peak_dB = 0.1
grating_coupler_loss_dB = 3
ng_noise = 0.1
length_variability = 1
for wafer in wafers:
for die in dies:
die = f"{(die['x'])}_{(die['y'])}"
for (_, row), (_, device_row) in zip(df.iterrows(), df.iterrows()):
cell_id = row["cell"]
if "ridge" in cell_id:
continue
top_cell_id = "rings"
device_id = f"{top_cell_id}_{cell_id}_{device_row['x']}_{device_row['y']}"
dirpath = cwd / wafer / die / device_id
dirpath.mkdir(exist_ok=True, parents=True)
data_file = dirpath / "data.json"
metadata_file = dirpath / "attributes.json"
metadata_file.write_text(json.dumps(metadata))
loss_dB = 2 * grating_coupler_loss_dB
peak_power = 10 ** (-loss_dB / 10)
grating_spectrum = np.array(
[
gaussian_grating_coupler_response(
peak_power, center_wavelength, bandwidth_1dB, wl
)
for wl in wavelength_range
]
)
ring_spectrum = ring(
wavelength_range,
wl0=1.55,
neff=2.4,
ng=4.0 + ng_noise * np.random.rand(),
ring_length=2 * np.pi * row["radius_um"]
+ length_variability * np.random.rand(),
coupling=0.1,
loss=30e2, # 30 dB/cm = 30000 dB/m
)
output_power = grating_spectrum * ring_spectrum
output_power *= 10 ** (
noise_peak_to_peak_dB * np.random.rand(len(wavelength_range)) / 10
)
d = {
"wavelength": wavelength_range * 1e3,
"output_power": output_power,
"polyfit": loss_dB,
}
data = pd.DataFrame(d)
json_converted_file = data.reset_index(drop=True).to_dict(orient="split")
json.dump(
json_converted_file,
open(data_file.with_suffix(".json"), "w+"),
indent=2,
)
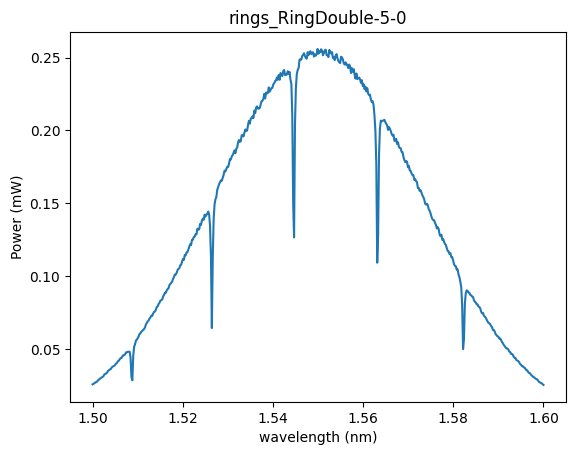
plt.plot(wavelength_range, output_power)
plt.title(dirpath.stem)
plt.ylabel("Power (mW)")
plt.xlabel("wavelength (nm)")
Text(0.5, 0, 'wavelength (nm)')
f"{len(list(dirpath.parent.glob('*/*.json')))//2} measurements"
'9 measurements'
f"{len(list(dirpath.parent.parent.glob('*')))} dies"
'64 dies'
